The examples in this document are taken from Singer and Willett (n.d.) and the translation into tidyverse and brms code by Kurz (2021).
We will first briefly look at how to reshape datasets, and then investigate two examples of longitudinal data with a hierarchical structure.
library(kableExtra)
library(tidyverse)
library(brms)
# set ggplot theme
theme_set(theme_grey(base_size = 14) +
theme(panel.grid = element_blank()))
# set rstan options
rstan::rstan_options(auto_write = TRUE)
options(mc.cores = 4)
Exploring data
We’ll load a dataset from RAUDENBUSH and CHAN (1992).
tolerance <- read_csv("https://stats.idre.ucla.edu/wp-content/uploads/2016/02/tolerance1.txt", col_names = TRUE)
head(tolerance, n = 16)
# A tibble: 16 x 8
id tol11 tol12 tol13 tol14 tol15 male exposure
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 9 2.23 1.79 1.9 2.12 2.66 0 1.54
2 45 1.12 1.45 1.45 1.45 1.99 1 1.16
3 268 1.45 1.34 1.99 1.79 1.34 1 0.9
4 314 1.22 1.22 1.55 1.12 1.12 0 0.81
5 442 1.45 1.99 1.45 1.67 1.9 0 1.13
6 514 1.34 1.67 2.23 2.12 2.44 1 0.9
7 569 1.79 1.9 1.9 1.99 1.99 0 1.99
8 624 1.12 1.12 1.22 1.12 1.22 1 0.98
9 723 1.22 1.34 1.12 1 1.12 0 0.81
10 918 1 1 1.22 1.99 1.22 0 1.21
11 949 1.99 1.55 1.12 1.45 1.55 1 0.93
12 978 1.22 1.34 2.12 3.46 3.32 1 1.59
13 1105 1.34 1.9 1.99 1.9 2.12 1 1.38
14 1542 1.22 1.22 1.99 1.79 2.12 0 1.44
15 1552 1 1.12 2.23 1.55 1.55 0 1.04
16 1653 1.11 1.11 1.34 1.55 2.12 0 1.25The data are in the wide format (one row per id), but we need one row per observation, so we need to rehape the data into a long format.
tolerance <- tolerance |>
pivot_longer(-c(id, male, exposure),
names_to = "age",
values_to = "tolerance") |>
# remove the `tol` prefix from the `age` values
mutate(age = str_remove(age, "tol") |> as.integer()) |>
arrange(id, age)
We have 16 subjects.
tolerance %>%
distinct(id) %>%
count()
# A tibble: 1 x 1
n
<int>
1 16tolerance %>%
slice(c(1:9, 76:80))
# A tibble: 14 x 5
id male exposure age tolerance
<dbl> <dbl> <dbl> <int> <dbl>
1 9 0 1.54 11 2.23
2 9 0 1.54 12 1.79
3 9 0 1.54 13 1.9
4 9 0 1.54 14 2.12
5 9 0 1.54 15 2.66
6 45 1 1.16 11 1.12
7 45 1 1.16 12 1.45
8 45 1 1.16 13 1.45
9 45 1 1.16 14 1.45
10 1653 0 1.25 11 1.11
11 1653 0 1.25 12 1.11
12 1653 0 1.25 13 1.34
13 1653 0 1.25 14 1.55
14 1653 0 1.25 15 2.12First, we will plot the observations over time (at different ages), and connect them with lines.
tolerance %>%
ggplot(aes(x = age, y = tolerance)) +
geom_point() +
geom_line() +
coord_cartesian(ylim = c(1, 4)) +
theme(panel.grid = element_blank()) +
facet_wrap(~id)
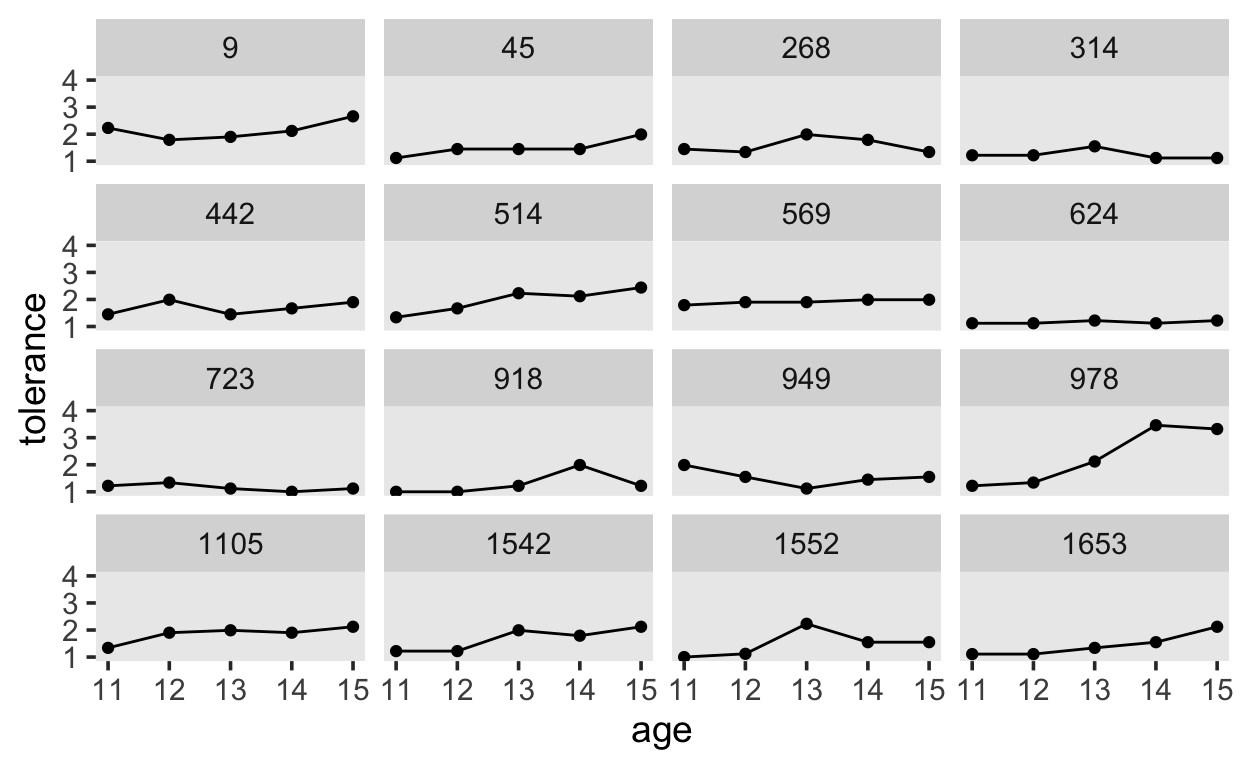
Next, we will apply locally weighted smoothing.
tolerance %>%
ggplot(aes(x = age, y = tolerance)) +
geom_point() +
stat_smooth(method = "loess", se = F, span = .9) +
coord_cartesian(ylim = c(1, 4)) +
theme(panel.grid = element_blank()) +
facet_wrap(~id)
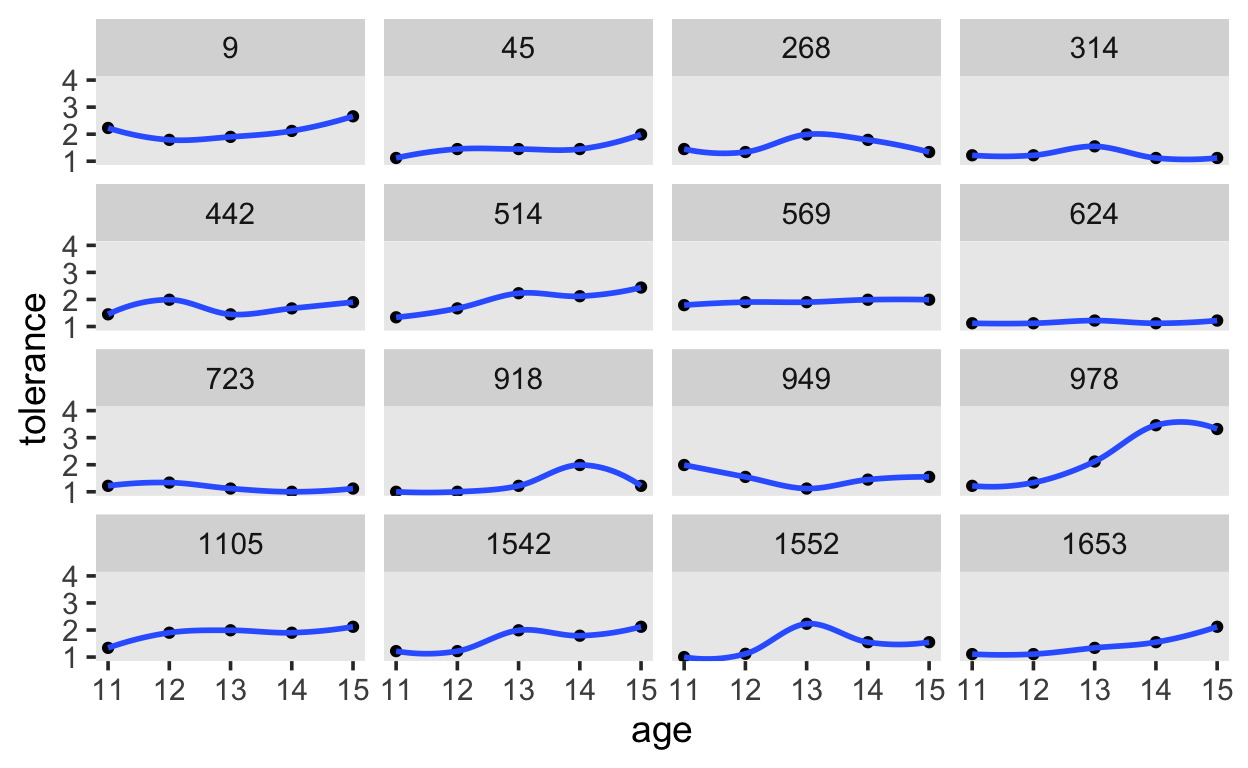
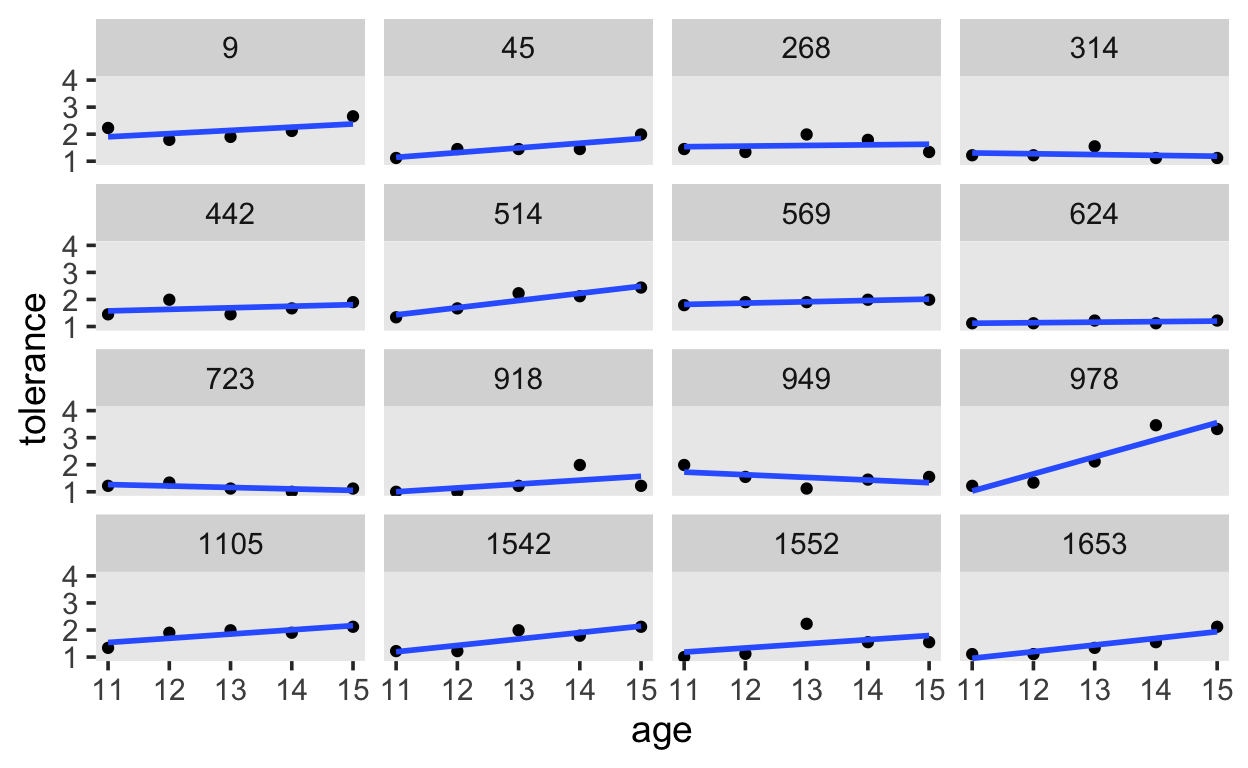
And finally, we’ll estimate the linear effect of age.
tolerance %>%
ggplot(aes(x = age, y = tolerance)) +
geom_point() +
stat_smooth(method = "lm", se = F, span = .9) +
coord_cartesian(ylim = c(1, 4)) +
theme(panel.grid = element_blank()) +
facet_wrap(~id)
Early Intervention
We’ll start with a data set on early intervention on child development (Singer and Willett n.d.)
As part of a larger study of the effects of early intervention on child development, these researchers tracked the cognitive performance of 103 African- American infants born into low-income families. When the children were 6 months old, approximately half (n = 58) were randomly assigned to participate in an intensive early intervention program designed to enhance their cognitive functioning; the other half (n = 45) received no intervention and constituted a control group. Each child was assessed 12 times between ages 6 and 96 months. Here, we examine the effects of program participation on changes in cognitive performance as measured by a nationally normed test administered three times, at ages 12, 18, and 24 months.
Each child has three records, one per wave of data collection. Each record contains four variables: (1) ID; (2) AGE, the child’s age (in years) at each assessment (1.0, 1.5, or 2.0); (3) COG, the child’s cognitive performance score at that age; and (4) PROGRAM, a dichotomy that describes whether the child participated in the early intervention program. Because children remained in their group for the duration of data collection, this predictor is time-invariant.
early_int <- read_csv("https://raw.githubusercontent.com/awellis/learnmultilevelmodels/main/data/early-intervention.csv") |>
mutate(id = as_factor(id),
intervention = factor(ifelse(program == 0, "no", "yes"),
levels = c("no", "yes")))
head(early_int, 10)
# A tibble: 10 x 6
id age cog program age_c intervention
<fct> <dbl> <dbl> <dbl> <dbl> <fct>
1 1 1 117 1 0 yes
2 1 1.5 113 1 0.5 yes
3 1 2 109 1 1 yes
4 2 1 108 1 0 yes
5 2 1.5 112 1 0.5 yes
6 2 2 102 1 1 yes
7 3 1 112 1 0 yes
8 3 1.5 113 1 0.5 yes
9 3 2 85 1 1 yes
10 4 1 138 1 0 yes In addition, we have the age-1 variable, age_c. This variable has the value 0 when the child is 1 year old.
early_int |>
filter(id %in% sample(levels(early_int$id), 12)) |>
ggplot(aes(x = age, y = cog)) +
stat_smooth(method = "lm", se = F) +
geom_point() +
scale_x_continuous(breaks = c(1, 1.5, 2)) +
ylim(50, 150) +
theme(panel.grid = element_blank()) +
facet_wrap(~id, ncol = 4)
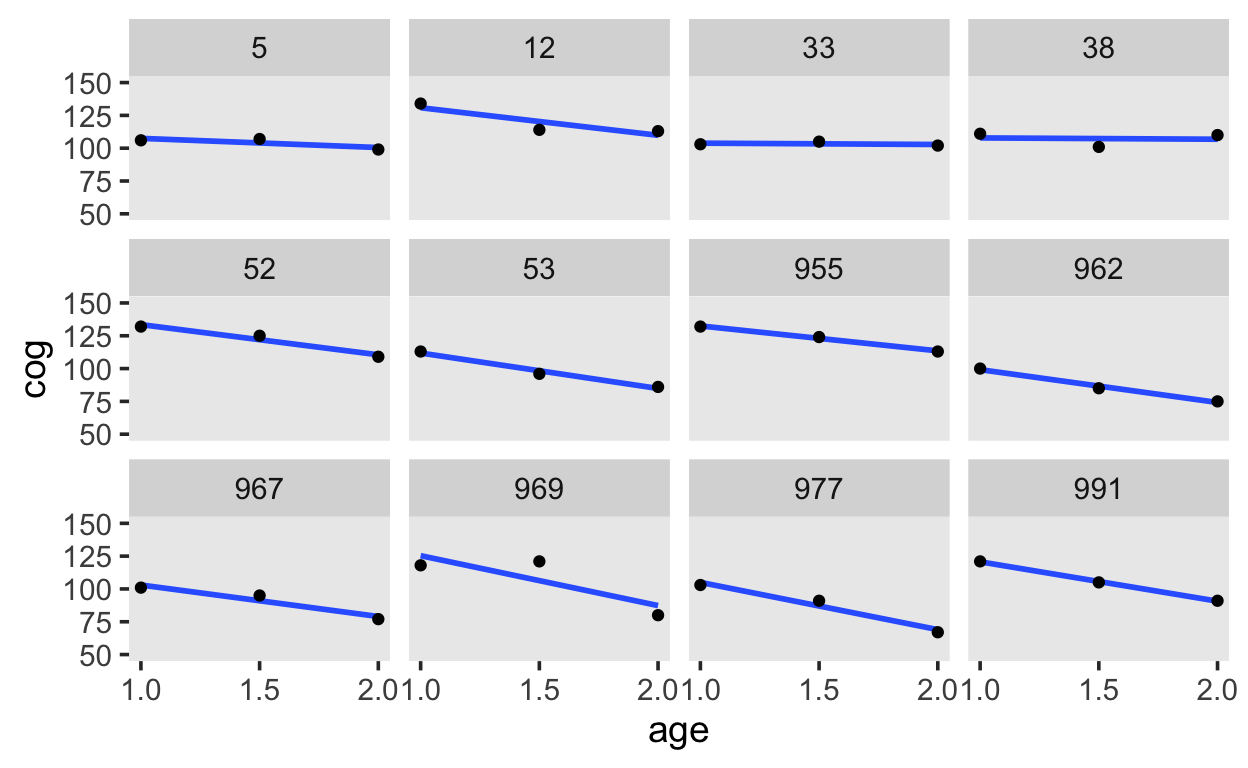
Our model for each child’s development is:
\[ \text{cog}_{ij} = [ \pi_{0i} + \pi_{1i} (\text{age}_{ij} - 1) ] + [\epsilon_{ij}]. \]
\[\begin{align*} \text{cog} & \sim \operatorname{Normal} (\mu_{ij}, \sigma_\epsilon^2) \\ \mu_{ij} & = \pi_{0i} + \pi_{1i} (\text{age}_{ij} - 1). \end{align*}\]
The \(i^{th}\) child’s cog score at observation \(j\) is normally distributed, with mean \(\mu\) and residuak standard deviation \(\sigma_{\epsilon}\). The mean \(\mu\) is modelled as a linear function of age. The intercept will correspond to the expected score at age 1.
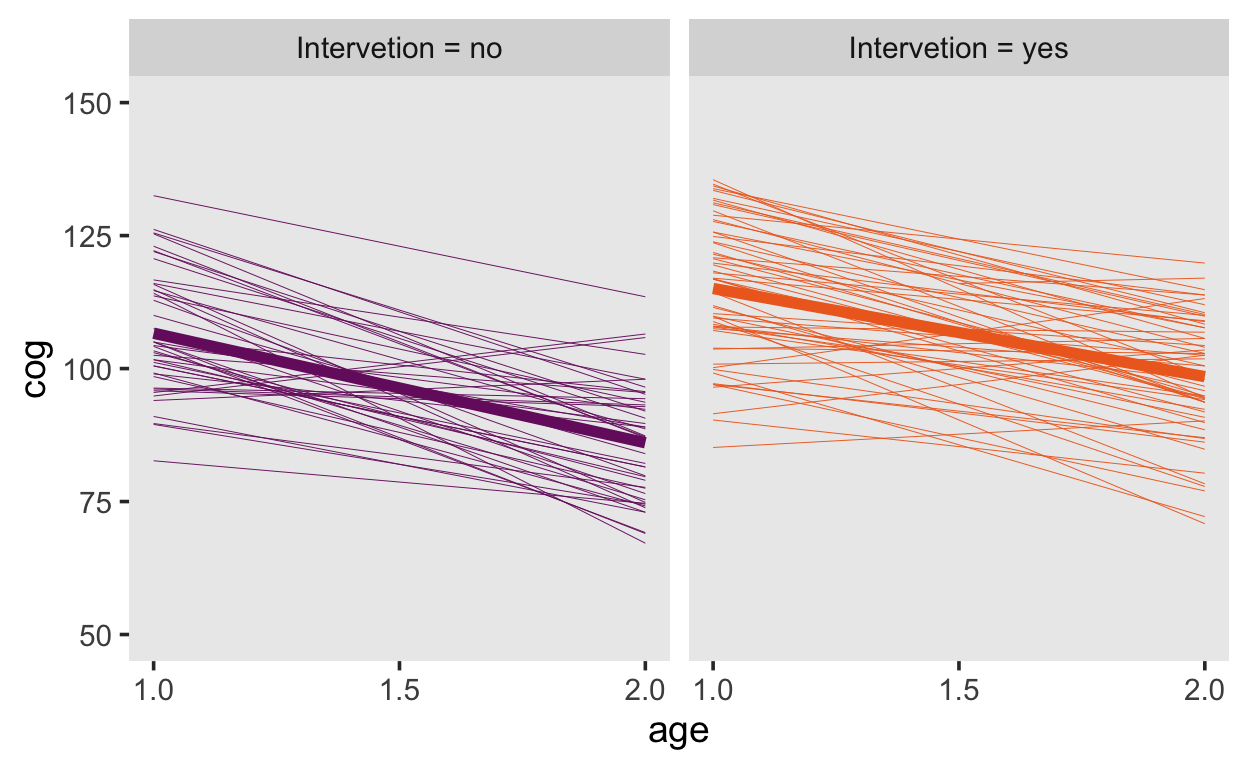
Next, we consider that the children are assigned to two different group.
early_int<-
early_int|>
mutate(label = str_c("Intervetion = ", intervention))
early_int |>
ggplot(aes(x = age, y = cog, color = label)) +
stat_smooth(aes(group = id),
method = "lm", se = F, size = 1/6) +
stat_smooth(method = "lm", se = F, size = 2) +
scale_color_viridis_d(option = "B", begin = .33, end = .67) +
scale_x_continuous(breaks = c(1, 1.5, 2)) +
ylim(50, 150) +
theme(legend.position = "none",
panel.grid = element_blank()) +
facet_wrap(~label)
Fitting the model with brms
fit_early_1
Family: gaussian
Links: mu = identity; sigma = identity
Formula: cog ~ 0 + Intercept + age_c + intervention + age_c:intervention + (1 + age_c | id)
Data: early_int (Number of observations: 309)
Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup samples = 4000
Group-Level Effects:
~id (Number of levels: 103)
Estimate Est.Error l-95% CI u-95% CI Rhat
sd(Intercept) 9.72 1.17 7.63 12.21 1.00
sd(age_c) 3.90 2.37 0.25 8.72 1.01
cor(Intercept,age_c) -0.48 0.35 -0.96 0.51 1.00
Bulk_ESS Tail_ESS
sd(Intercept) 830 2186
sd(age_c) 274 498
cor(Intercept,age_c) 2951 1938
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat
Intercept 105.72 1.86 102.02 109.32 1.00
age_c -19.92 1.90 -23.66 -16.21 1.00
interventionyes 9.79 2.41 4.91 14.61 1.00
age_c:interventionyes 2.66 2.50 -2.27 7.59 1.00
Bulk_ESS Tail_ESS
Intercept 1247 1818
age_c 2249 2811
interventionyes 1338 2137
age_c:interventionyes 2395 2714
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 8.59 0.50 7.63 9.59 1.01 513 1347
Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).conditional_effects(fit_early_1,
"age_c:intervention")
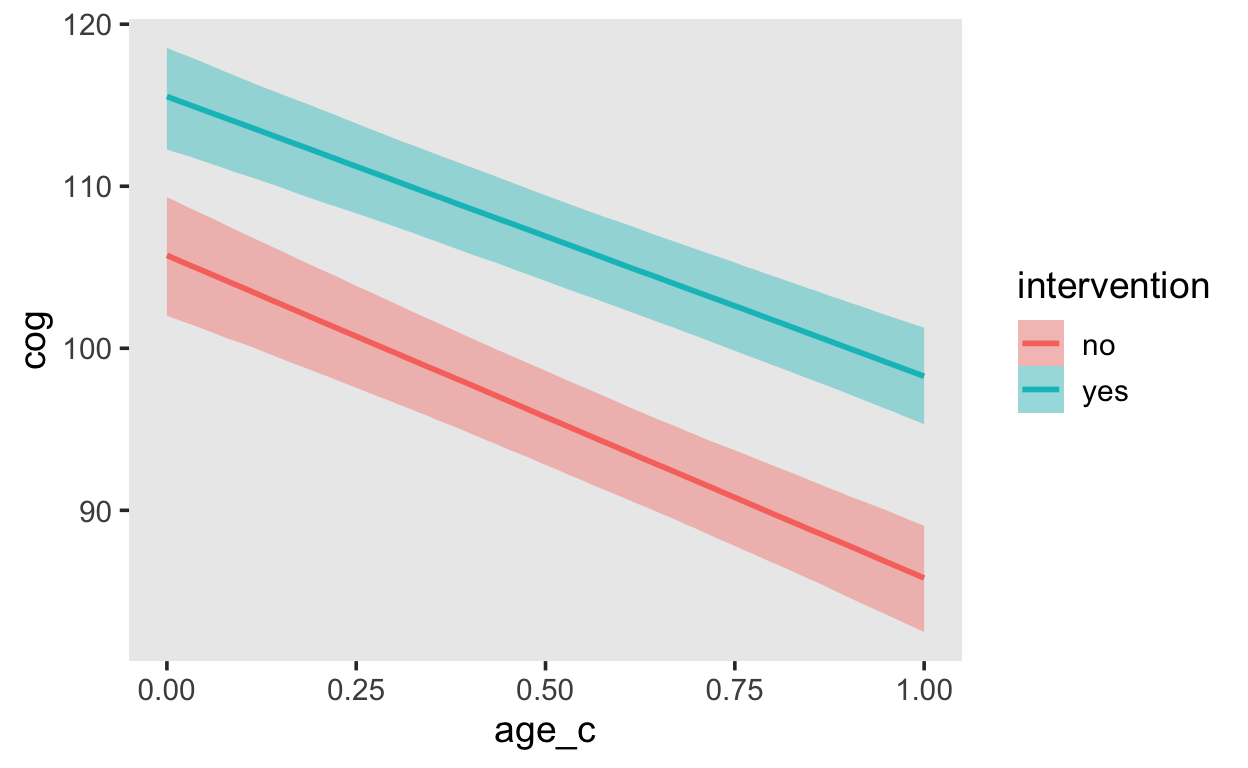
How would you go about demonstrating evidence for or against a difference between the interventions?
Changes in adolescent alcohol use
As part of a larger study of substance abuse, Curran, Stice, and Chassin (1997) collected three waves of longitudinal data on 82 adolescents. Each year, beginning at age 14, the teenagers completed a four-item instrument assessing their alcohol consumption during the previous year. Using an 8-point scale (ranging from 0 = “not at all” to 7 = “every day”), adolescents described the frequency with which they (1) drank beer or wine, (2) drank hard liquor, (3) had five or more drinks in a row, and (4) got drunk. The data set also includes two potential predictors of alcohol use: COA, a dichotomy indicating whether the adolescent is a child of an alcoholic parent; and PEER, a measure of alcohol use among the adolescent’s peers. This latter predictor was based on information gathered during the initial wave of data collection. Participants used a 6-point scale (ranging from 0 = “none” to 5 = “all”) to estimate the proportion of their friends who drank alcohol occasionally (one item) or regularly (a second item).
Do individual trajectories of alcohol use during adolescence differ according to the history of parental alcoholism and early peer alcohol use.
library(tidyverse)
alcohol <- read_csv("https://raw.githubusercontent.com/awellis/learnmultilevelmodels/main/data/alcohol1_pp.csv")
head(alcohol)
# A tibble: 6 x 9
id age coa male age_14 alcuse peer cpeer ccoa
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 14 1 0 0 1.73 1.26 0.247 0.549
2 1 15 1 0 1 2 1.26 0.247 0.549
3 1 16 1 0 2 2 1.26 0.247 0.549
4 2 14 1 1 0 0 0.894 -0.124 0.549
5 2 15 1 1 1 0 0.894 -0.124 0.549
6 2 16 1 1 2 1 0.894 -0.124 0.549| Variable | Description |
|---|---|
| id | subject ID |
| age | in years |
| coa | child of alcoholic parent (1 - yes, 0 - no) |
| male | indicator for male |
| age_14 | age - 14 (0 corresponds to age: 14) |
| alcuse | alcohol consumption |
| peer | alcohol use among peers |
| cpeer, ccoa | centred variables |
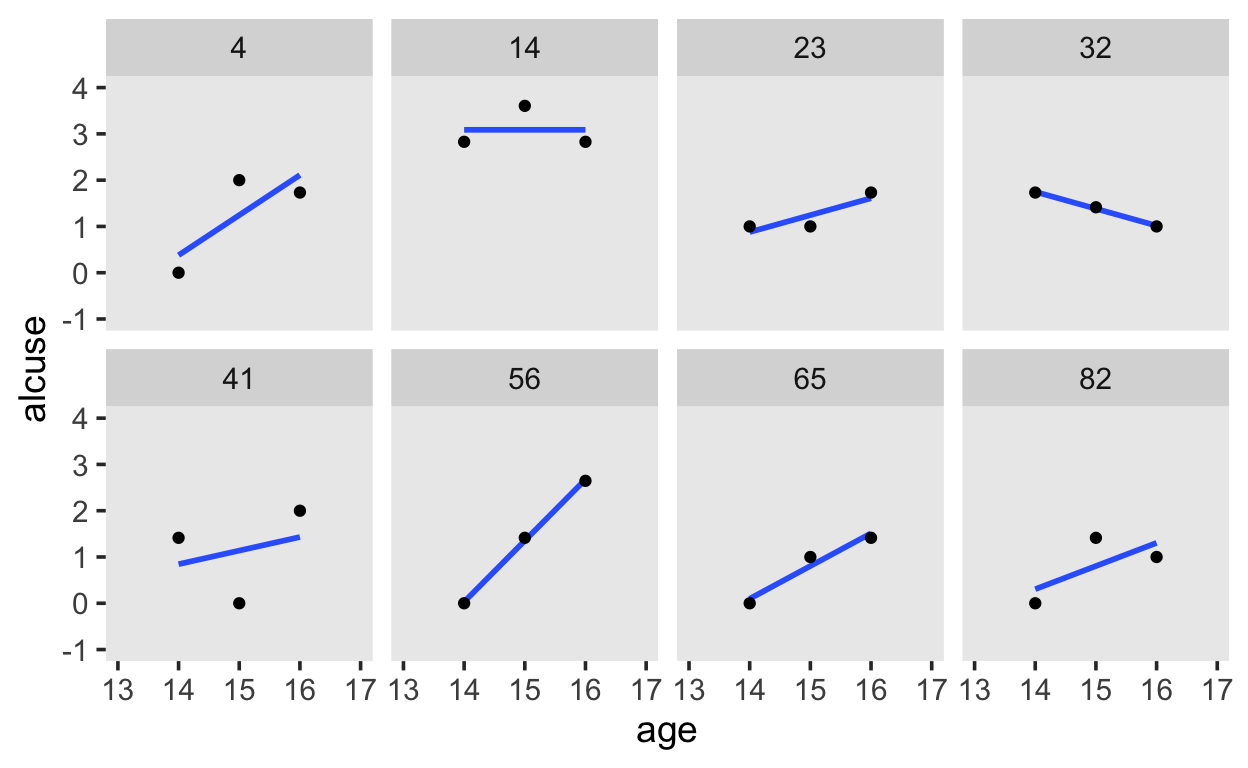
alcohol %>%
filter(id %in% c(4, 14, 23, 32, 41, 56, 65, 82)) %>%
ggplot(aes(x = age, y = alcuse)) +
stat_smooth(method = "lm", se = F) +
geom_point() +
coord_cartesian(xlim = c(13, 17),
ylim = c(-1, 4)) +
theme(panel.grid = element_blank()) +
facet_wrap(~id, ncol = 4)
\[\begin{align*} \text{alcuse}_{ij} & = \pi_{0i} + \pi_{1i} (\text{age}_{ij} - 14) + \epsilon_{ij}\\ \epsilon_{ij} & \sim \operatorname{Normal}(0, \sigma_\epsilon^2), \end{align*}\]
\[\begin{align*} \text{alcuse}_{ij} & = \big [ \gamma_{00} + \gamma_{10} \text{age_14}_{ij} + \gamma_{01} \text{coa}_i + \gamma_{11} (\text{coa}_i \times \text{age_14}_{ij}) \big ] \\ & \;\;\;\;\; + [ \zeta_{0i} + \zeta_{1i} \text{age_14}_{ij} + \epsilon_{ij} ] \\ \epsilon_{ij} & \sim \operatorname{Normal} (0, \sigma_\epsilon^2) \\ \begin{bmatrix} \zeta_{0i} \\ \zeta_{1i} \end{bmatrix} & \sim \operatorname{Normal} \begin{pmatrix} \begin{bmatrix} 0 \\ 0 \end{bmatrix}, \begin{bmatrix} \sigma_0^2 & \sigma_{01} \\ \sigma_{01} & \sigma_1^2 \end{bmatrix} \end{pmatrix}, \end{align*}\]
Unconditional means model
\[\begin{align*} \text{alcuse}_{ij} & = \gamma_{00} + \zeta_{0i} + \epsilon_{ij} \\ \epsilon_{ij} & \sim \operatorname{Normal}(0, \sigma_\epsilon^2) \\ \zeta_{0i} & \sim \operatorname{Normal}(0, \sigma_0^2). \end{align*}\]
get_prior(alcuse ~ 1 + (1 | id),
data = alcohol)
prior class coef group resp dpar nlpar bound
student_t(3, 1, 2.5) Intercept
student_t(3, 0, 2.5) sd
student_t(3, 0, 2.5) sd id
student_t(3, 0, 2.5) sd Intercept id
student_t(3, 0, 2.5) sigma
source
default
default
(vectorized)
(vectorized)
defaultfit_alcuse_1 <-
brm(alcuse ~ 1 + (1 | id),
data = alcohol,
file = "models/fit_alcuse_1",
file_refit = "on_change")
Unconditional growth model
\[\begin{align*} \text{alcuse}_{ij} & = \gamma_{00} + \gamma_{10} \text{age_14}_{ij} + \zeta_{0i} + \zeta_{1i} \text{age_14}_{ij} + \epsilon_{ij} \\ \epsilon_{ij} & \sim \operatorname{Normal} (0, \sigma_\epsilon^2) \\ \begin{bmatrix} \zeta_{0i} \\ \zeta_{1i} \end{bmatrix} & \sim \operatorname{Normal} \begin{pmatrix} \begin{bmatrix} 0 \\ 0 \end{bmatrix}, \begin{bmatrix} \sigma_0^2 & \sigma_{01} \\ \sigma_{01} & \sigma_1^2 \end{bmatrix} \end{pmatrix}. \end{align*}\]
get_prior(alcuse ~ 0 + Intercept + age_14 + (1 + age_14 | id),
data = alcohol)
prior class coef group resp dpar nlpar bound
(flat) b
(flat) b age_14
(flat) b Intercept
lkj(1) cor
lkj(1) cor id
student_t(3, 0, 2.5) sd
student_t(3, 0, 2.5) sd id
student_t(3, 0, 2.5) sd age_14 id
student_t(3, 0, 2.5) sd Intercept id
student_t(3, 0, 2.5) sigma
source
default
(vectorized)
(vectorized)
default
(vectorized)
default
(vectorized)
(vectorized)
(vectorized)
defaultEffect of COA
\[\begin{align*} \text{alcuse}_{ij} & = \gamma_{00} + \gamma_{01} \text{coa}_i + \gamma_{10} \text{age_14}_{ij} + \gamma_{11} \text{coa}_i \times \text{age_14}_{ij} + \zeta_{0i} + \zeta_{1i} \text{age_14}_{ij} + \epsilon_{ij} \\ \epsilon_{ij} & \sim \text{Normal} (0, \sigma_\epsilon^2) \\ \begin{bmatrix} \zeta_{0i} \\ \zeta_{1i} \end{bmatrix} & \sim \text{Normal} \begin{pmatrix} \begin{bmatrix} 0 \\ 0 \end{bmatrix}, \begin{bmatrix} \sigma_0^2 & \sigma_{01} \\ \sigma_{01} & \sigma_1^2 \end{bmatrix} \end{pmatrix}. \end{align*}\]
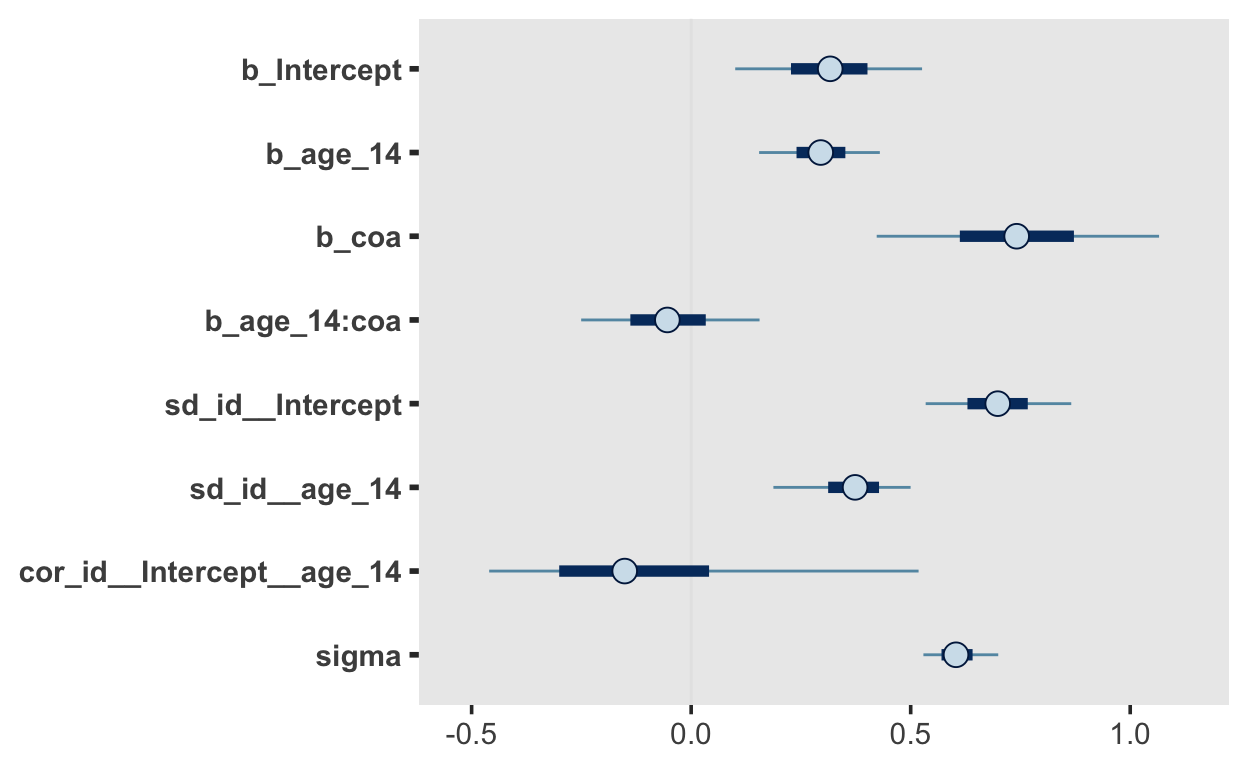
fit_alcuse_3
Family: gaussian
Links: mu = identity; sigma = identity
Formula: alcuse ~ 0 + Intercept + age_14 + coa + age_14:coa + (1 + age_14 | id)
Data: alcohol (Number of observations: 246)
Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup samples = 4000
Group-Level Effects:
~id (Number of levels: 82)
Estimate Est.Error l-95% CI u-95% CI Rhat
sd(Intercept) 0.70 0.10 0.51 0.90 1.01
sd(age_14) 0.36 0.09 0.15 0.52 1.02
cor(Intercept,age_14) -0.10 0.29 -0.50 0.70 1.01
Bulk_ESS Tail_ESS
sd(Intercept) 947 2048
sd(age_14) 341 668
cor(Intercept,age_14) 624 601
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
Intercept 0.32 0.13 0.06 0.57 1.00 2454
age_14 0.29 0.08 0.13 0.46 1.00 3114
coa 0.74 0.19 0.36 1.12 1.00 2196
age_14:coa -0.05 0.12 -0.29 0.19 1.00 2956
Tail_ESS
Intercept 2757
age_14 2854
coa 2481
age_14:coa 2883
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.61 0.05 0.52 0.72 1.01 524 1151
Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).mcmc_plot(fit_alcuse_3)
loo_compare(loo_alcuse_2, loo_alcuse_3)
elpd_diff se_diff
fit_alcuse_2 0.0 0.0
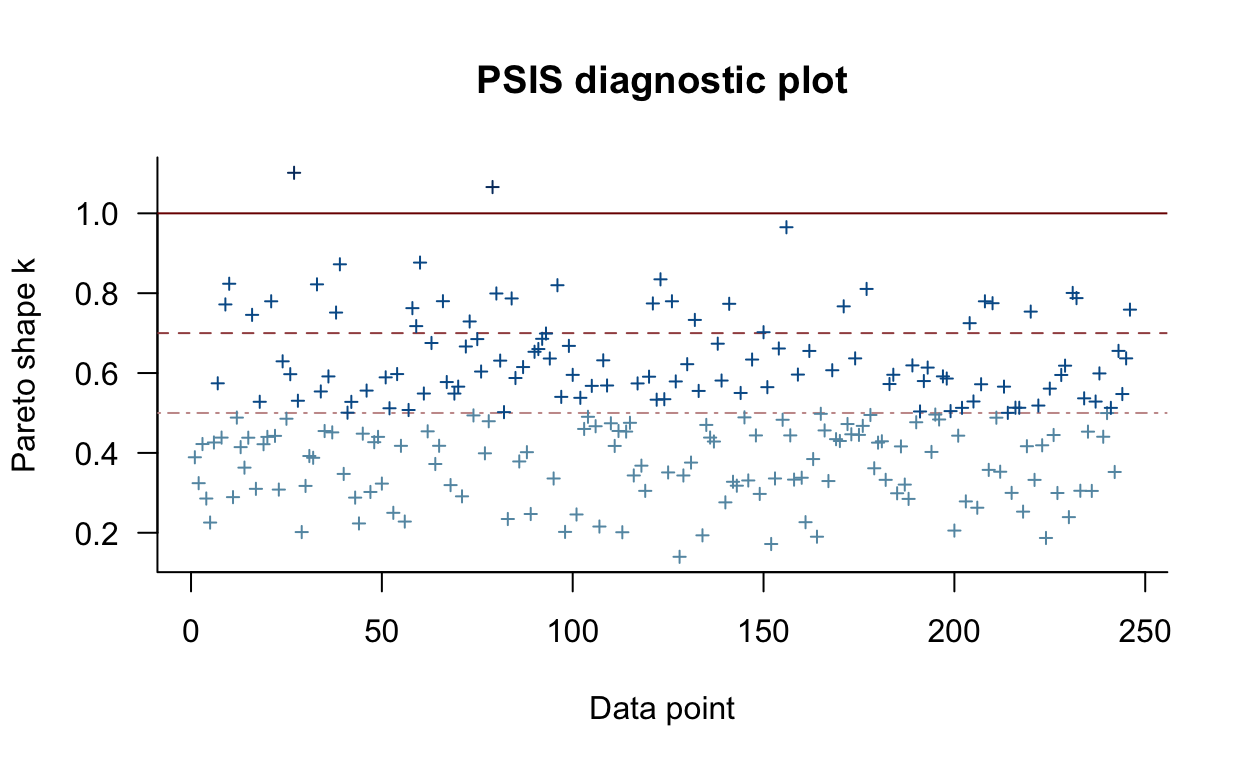
fit_alcuse_3 -0.7 2.4 plot(loo_alcuse_3)
Time-varying predictors
If we have time-varying predictor variable, it is a good idea the decompose these into a subject’specific mean (trait) and a session-by-session deviation from that mean (state).
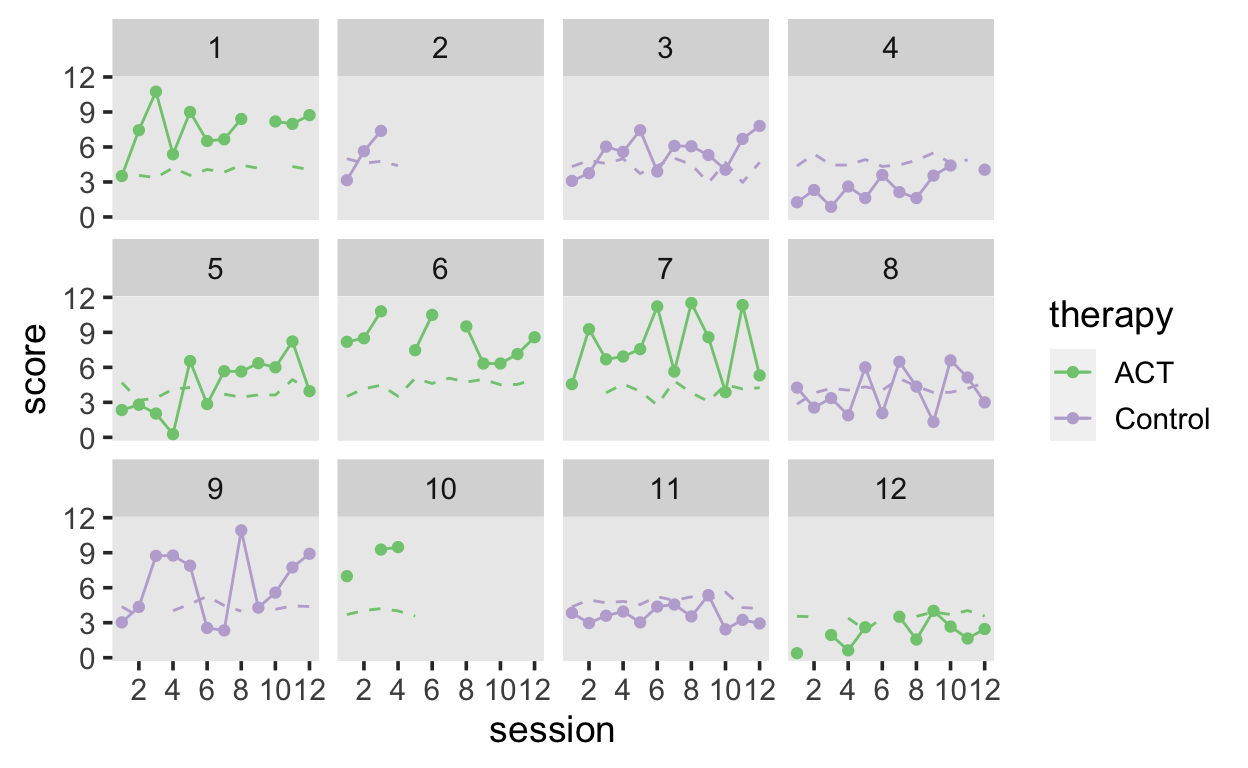
Let’s look at some simulated data, in which patient are assigned to either a control or a therapy (ACT) group. Patients’ wellbeing (score) is assessed over the course of 12 sessions. In addition, we have the patient-rated therapeutic alliance, which reflects how the quality of patient-therapist interactions.
wellbeing <- read_csv("https://raw.githubusercontent.com/awellis/learnmultilevelmodels/main/data/wellbeing.csv")
wellbeing |>
ggplot(aes(session, score, color = therapy)) +
geom_line() +
geom_point() +
geom_line(aes(session, alliance), linetype = 2) +
scale_x_continuous(breaks = seq(2, 12, by = 2)) +
scale_color_brewer(type = "qual") +
facet_wrap(~patient)
We want to predict patient’s wellbeing as a function of the therapeutic alliance, while accounting for a linear trend.
- How would you specify this model, as well as unconditional models?
- At which level is the predictor variable
alliance?
Decomposing time-varying predictors
Instead of using the raw alliance variable, we will create two new variables: the average alliance, and the session-by-session deviations from the average. The average will be a patient-level (level 2) predictor, and can be used to explain variability between patients, whereas the deviations reflect fluctuations within patients at individual sessions.
fit_alliance_1 <- brm(score ~ session + (1 + session| patient),
data = wellbeing,
file = "models/fit_alliance_1")
fit_alliance_2 <- brm(score ~ session + al_between + al_within +
(1 + session + al_within | patient),
data = wellbeing,
file = "models/fit_alliance_2")
fit_alliance_3 <- brm(score ~ session + al_within +
(1 + session + al_within | patient),
data = wellbeing,
file = "models/fit_alliance_3")
fit_alliance_2
Family: gaussian
Links: mu = identity; sigma = identity
Formula: score ~ session + al_between + al_within + (1 + session + al_within | patient)
Data: wellbeing (Number of observations: 111)
Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup samples = 4000
Group-Level Effects:
~patient (Number of levels: 12)
Estimate Est.Error l-95% CI u-95% CI Rhat
sd(Intercept) 2.67 0.73 1.58 4.34 1.00
sd(session) 0.09 0.07 0.00 0.27 1.00
sd(al_within) 0.90 0.57 0.05 2.21 1.00
cor(Intercept,session) -0.14 0.46 -0.87 0.80 1.00
cor(Intercept,al_within) -0.37 0.44 -0.96 0.67 1.00
cor(session,al_within) -0.11 0.49 -0.89 0.84 1.00
Bulk_ESS Tail_ESS
sd(Intercept) 1566 2500
sd(session) 1304 1738
sd(al_within) 1661 1780
cor(Intercept,session) 2799 2463
cor(Intercept,al_within) 2895 2439
cor(session,al_within) 2263 3065
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
Intercept 4.89 8.00 -10.44 21.29 1.00 1426
session 0.18 0.06 0.05 0.30 1.00 3139
al_between -0.11 1.88 -3.94 3.54 1.00 1440
al_within -0.72 0.50 -1.72 0.22 1.00 2778
Tail_ESS
Intercept 1882
session 2514
al_between 1880
al_within 2668
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 1.80 0.14 1.55 2.10 1.00 3490 2939
Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).